Systematically Uncovering the Hidden Biochemical Activities within Human Transcription Factors
Transcription factors lie at the heart of gene regulation, serving as molecular switches that control cellular fates and developmental outcomes. Despite decades of research, we still lack a systematic framework for understanding how transcription factors regulate gene expression in human cells. This gap has profound consequences: although the human genome is densely annotated with transcription factor binding sites, we still cannot predict what most of those sites do. Transcriptional output is ultimately shaped by a complex network of molecular interactions that occur beyond DNA binding. These interactions are often mediated by regions located within the unstructured regions of transcription factors, regions that, despite their critical regulatory roles, have been historically overlooked and technically challenging to study. There is an urgent need for new approaches capable of capturing these transient interactions at scale.
To address these challenges, our lab uniquely unites two historically separate approaches: high-throughput profiling of molecular activities inside living cells and precise, quantitative measurements collected in vitro. We do this using variants of a high-throughput recruitement assay, HT-recruit, and a high-throughput microfluidic binding assay, STAMMPPING. By integrating synthetic biology, microfluidics, and biophysics with scalable assays, we can seamlessly toggle between molecules and cells, building quantitative, bottom-up models that link molecular interactions to transcription factor function. This approach enables us to map the nodes and connections within the transcription factor interaction network at unprecedented scale, build predictive models of human gene regulation and, ultimately, develop new strategies for engineering cellular behavior and targeting these regulatory interactions.
-
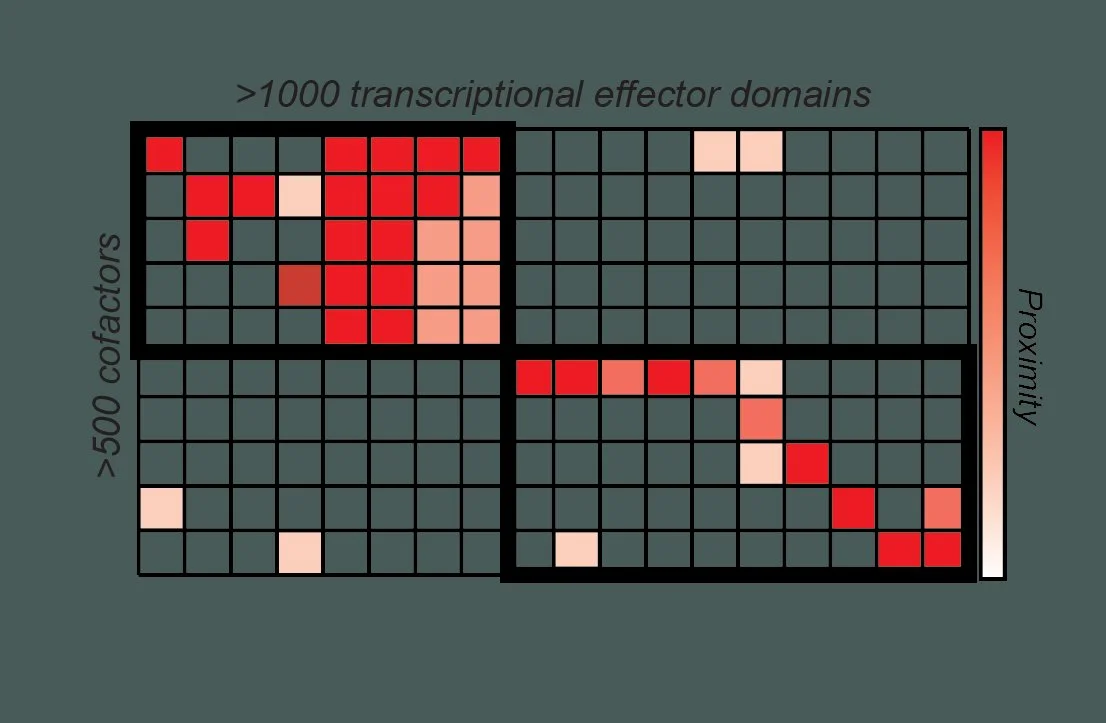
Who binds whom during transcription?
Human genomes are regulated by a complex network of protein interactions, where intrinsically disordered regions within transcription factors interact with well-folded domains in chromatin regulators, co-activators, and co-repressors to modulate RNA Polymerase II activity. However, the vast majority of these interactions are unknown. We are developing new high-throughput approaches for systematically mapping these interaction networks across cell types to uncover general principles of transcription factor-mediated gene regulation.
-

How do binding interactions dictate the strength and timing of gene regulation?
While we know transcription factors engage cofactors to drive gene expression, we still lack a predictive framework for how the biochemical properties of individual interactions—such as binding affinity, kinetics, and specificity—translate into functional transcriptional outputs. We are uniting two historically separate techniques: high-throughput profiling of molecular activities inside living cells and precise, quantitative measurements collected in vitro in order to build input/output models of transcription.
-
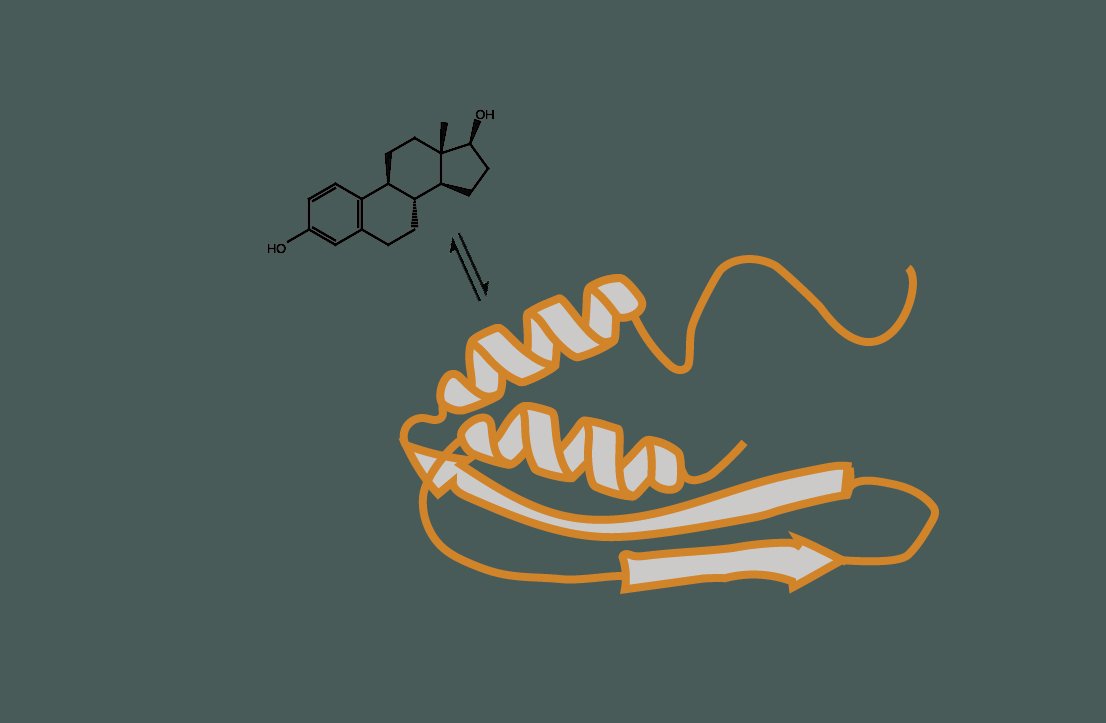
How many transcriptional proteins sense and respond to signaling molecules?
Signaling molecules, such as hormones, growth factors, and other extracellular signals, bind to receptors on the cell surface, and initiate intracellular signaling cascades that ultimately lead to the activation or repression of specific genes. Transcriptional proteins are crucial in interpreting these signals and regulating gene expression in response. We are developing new high-throughput assays to systematically identify proteins that act as both signal receivers and regulators of gene expression.